(Last updated: 11/07/2012)
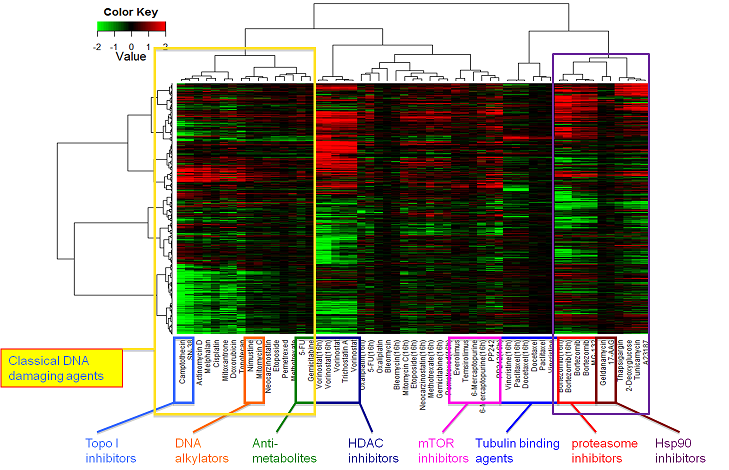
Human colon adenocarcinoma HT-29 cells were treated with anticancer compounds at various concentrations for 6 or 16 h. Gene expression changes were analyzed with GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix).
Hierarchical cluster analysis based on the collection of gene signatures of 35 anticancer compounds (55 treatment samples). In total 3237 probe sets were used for clustering. The values in the heatmap are the logarithm of sample-to-control ratio of intensity values. Neither normalizing nor scaling was performed. Up- and down-regulated genes are colored in red and green, respectively.
Up- or down-regulated genes in the clusters.
| Mode of action | up-regulated | down-regulated |
|---|---|---|
| Classical DNA damaging agents (16 compounds) |
DNAdamage_up.csv | DNAdamage_down.csv |
| HDAC inhibitors (5) | HDACi_up.csv | HDACi_down.csv |
| mTOR inhibitors (5) | mTORi_up.csv | mTORi_down.csv |
| Tubulin binding agents (6) | Tubulin_up.csv | Tubulin_down.csv |
| proteasome inhibitors (5) | proteasomei_up.csv | proteasomei_down.csv |
| ER stress inducers (4) | ERstress_up.csv | ERstress_down.csv |
Based on our gene expression data of anticancer agents and the C-map algorithm (Lamb et al. 2006), we provide this calculation program (connectivity scoring analysis) to compare gene signatures of test compounds to those of antitumor agents in our database for prediction of their likely modes of action.
Example of input files: GR033-up3-300.txt (up) GR033-down3-300.txt (down)
Sorry, only a source code of R program is available now.
GUI version is under construction.
Data and program for download:
Usage:
> source("Cscore_analysis.R")
> cscore_analysis(file.up, file.down, file.out)
Input file format (file.up, file.down):
List of the probeset IDs of Affymetrix HG-U133Plus2.0 array in one column
This program depends on the BioConductor packages as follows:
Program for download:
Usage:
> source("KEGG_analysis.R")
> KEGG_analysis(file.in, file.out)
Input file format:
List of the probeset IDs of Affymetrix HG-U133Plus2.0 array in one column